User:Davisb/EMSegmentModule: Difference between revisions
| Line 196: | Line 196: | ||
=== Step 8: Segmentation === | === Step 8: Segmentation === | ||
*The user can save the template before or after running the algorithm | |||
* | *Run Algorithm | ||
*Progress bar | *Progress bar | ||
*User can interrupt execution | *User can interrupt execution | ||
*At completion the user can visualize segmentations as overlays on the loaded image volumes | *At completion the user can | ||
* | ** visualize segmentations as overlays on the loaded image volumes | ||
** press button to remove small islands | |||
** press button to generate 3D modeles | |||
==Description of MRML Nodes== | ==Description of MRML Nodes== | ||
Revision as of 21:35, 10 November 2006
Project Summary
The goal of this project is to implement a Slicer3 module which creates templates that will be used in segmentation tasks. A template is composed of atlas data and a non-trivial collection of parameters for the EM segmentation algorithm. The template, which is written as an XML file, can be used later to segment new images. The target audience for this module is someone familiar with brain atlases and tissue labels, not a computer scientist.
This module will be demonstrated at the NAMIC All-Hands meeting in January 2007.
Contacts
- MIT: Polina Golland, Kilian Pohl (pohl@csail.mit.edu)
- Kitware: Brad Davis (brad.davis@kitware.com), Will Schroeder (will.schroeder@kitware.com)
General Module Description
The purpose of the module is to build a template that can be used later to segment new images. The template is composed of atlas data and a non-trivial collection of parameters for the EM segmentation algorithm. It is the user's job to specify the parameters so that the template is effective for segmenting a particular target image pair (T1 and T2, e.g., from a particular scanner). Once the parameters are specified, the target images are segmented using the EM segmentation algorithm. If the results are satisfactory, the template is saved and can be used to segment new target images. If the results are not satisfactory, the parameters can be modified and the segmentation re-run.
The template building process is divided in to a sequence of steps in order to simplify template specification for the non-expert user.
What are the steps in the template building process?
- Anatomical Structure Hierarchy: Define a hierarchy of anatomical structures.
- Spatial Priors: Assign uniform priors or probability images for each leaf node of the hierarchy.
- Intensity Images: Assign target intensity images.
- Intensity Distributions: Define an intensity distribution for each leaf node of the hierarchy.
- Node-based Parameters: Specify node-based segmentation parameters.
- Global Parameters: Specify global segmentation parameters.
- Registration Parameters: Specify atlas-to-template registration parameters.
- Segmentation: Apply EM algorithm to segment target image pair.
How will the template be used?
The user will (1) load the template, (2) load a new, unsegmented target image pair (T1 and T2), (3) tweak parameters from the template if desired, and (4) start the segmentation process. If the input images are not aligned, they will be aligned prior to the segmentation process---this will be specified by the user.
Alternately, the template can be used by a batch process to segment a collection of images.
Will this process (applying the template) require any parameters that are not included in the template?
Yes. Parameters pertaining to the target images are not stored in the template. Specifically, the template does not store the (1) target volume IDs and (2) whether or not the target images must be aligned. All other parameters should be specified in the template.
Will the template building module contain parameters that are not included in the template?
Yes. The template will not contain information regarding the target images. Also, the template will not contain intermediate data used to compute parameters for the template (e.g., intensity samples used to compute mean and covariance).
Detailed Description of Wizard Steps
Step 1: Anatomical Structure Hierarchy
Build and edit a tree anatomical structures:
- add new structure (specify name & color)
- rename node, change associated color
- remove a node
- reorganize a node (e.g., specify a new parent for this node)
Step 2: Spatial Priors
For each leaf node specify a spatial prior by one of:
- Specify a uniform prior (double in [0,1])
- Specify a prior probability map (volume)
- Number of Training Samples for prior probability map ---int
Step 3: Intensity Images
- Specify atlas T1 and T2 volumes
- Specify target T1 and T2 volumes
- Should the target images be aligned prior to segmentation? Specify with a check-box.
Step 4: Intensity Distributions
For each leaf node specify mean and covariance of bivariate Gaussian distribution:
- parameters may be entered and edited manually
- parameters can be learned by clicking on a representative sample of voxels in the target intensity volumes (~8 clicks)
- the user may erase a particular sample or all samples
- A display will be available to visualize the intensity distributions
- plot of distributions for T1 and T2 intensities separately (marginal distributions, intensity vs. probability)
- 2D image visualization of probabilities for T1xT2 intensity combinations (T1 intensity vs. T2 intensity)
- saturation is proportional to probability
- In all plots, color is based on tissue type (color selected for leaf node in tree)
- The user can toggle the display of each tissue type on and off
- When the user mouses over the MR image, the intensity under the mouse is plotted on each graph
Image of Slicer2.6 visualization window
Step 5: Node-Based Parameters
For all nodes
- Class Probability---double
- Local Prior Weight---double
- Input Channel Weights---double for each input channel, final representation is char*
- Print flags
- Weights
For leaf nodes only
- Intensity mean and covariance: this is specified in previous step
- Print flags
- PCA
- Quality
- Requires specification of reference standard volume
For parent nodes only
- Frequency---int---used by some of the print flags
- Print flags
- Registration Parameters
- Registration Similarity Measure
- Bias---uses Frequency
- Label-map---uses Frequency
- Shape Similarity Measure
- EMLabelMapConvergence
- EMWeightsConvergence
- MFALabelMapConvergence
- MFAWeightsConvergence
- Stopping Condition Parameters
- EM Type---int
- EM Value---double
- EM Max Iterations---int
- MFA Type---int
- MFA Value---double
- MFA Max Iterations---int
- Bias Calculation---int
- Miscellaneous Parameters
- Local Prior Name---char*
- Registration Translation---double[3]
- Registration Rotation---double[3]
- Registration Scale---double[3]
- Registration Covariance---double[9]
- Registration Class Specific Registration Flag
- Exclude From Incomplete E-Step Flag
- Registration Type---int
- Generate Background Probability Flag
- PCA Shape Model Type---int
- Registration Independent Sub-class Flag
- Class Interaction Matrices
- 6 matrices: North, South, East, West, Up, Down; choose from list
- Each matrix is of size (number of subclasses) by (number of subclasses)
- Each element is a double
- The final representation for each matrix is char*
Step 6: Global Parameters
- Alpha---double
- Smoothing Kernel Width---int
- Smoothing Kernel Sigma---double
- Multi-threading Enabled---on/off
- Working Directory---char*
Non-template options:
- Run Remotely---on/off
- Remote Server---char*---should be something like davisb@swan.cs.unc.edu
Step 7: Registration Parameters
What type of registration should be run: a pull down list.
- Affine
- MI
- ....
- disabled
- non rigid
- Bspline
- AG
- ....
- disabled
- joint registration & segmentation
Choose interpolation type from pull down list
Alignment of input channels on/off
Step 8: Segmentation
- The user can save the template before or after running the algorithm
- Run Algorithm
- Progress bar
- User can interrupt execution
- At completion the user can
- visualize segmentations as overlays on the loaded image volumes
- press button to remove small islands
- press button to generate 3D modeles
Description of MRML Nodes
Overview
By writing out a template we just mean writing the MRML tree.
The template will include atlas data (i.e. specification of image files), anatomical structure hierarchy information, algorithm parameters, and display parameters (e.g., the graph color for a particular structure). The data, hierarchy, parameter, and display information will be defined in separate nodes. Dependencies and hierarchical structure will be maintained by references: for example, a leaf node will contain a reference to the ID of its parent node. This is the idiom that is used, for example, to nest transforms and associate image volumes with volume display parameters in Slicer3.
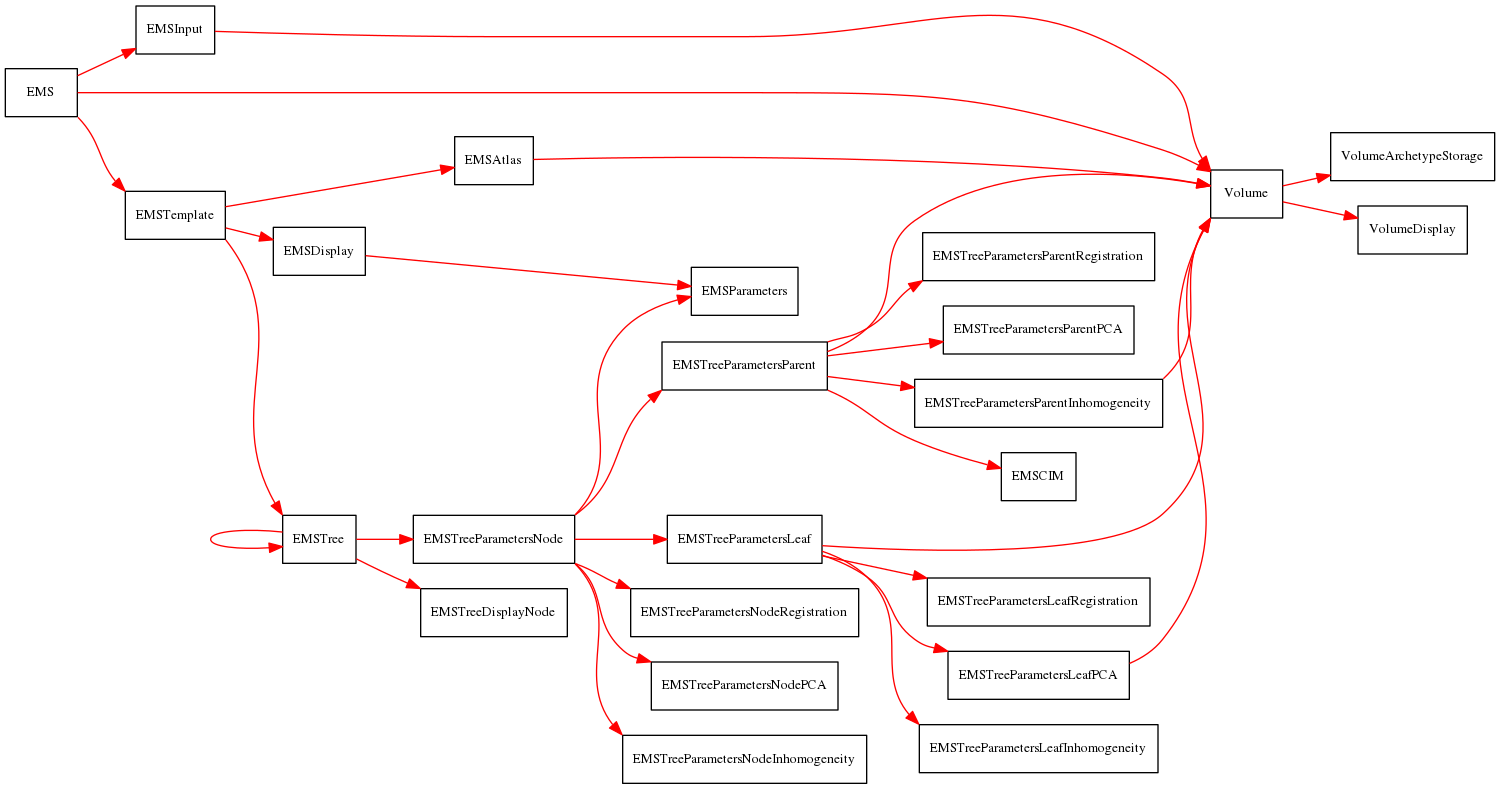
The following graph shows dependency information for all of the node types proposed for this file format. An arrow pointing from node A to node B indicates that (1) the XML tag for each node of type A contains a reference to the ID of a node of type B, and (2) the MRML node class representing an A node contains a pointer to a MRML B node class.
The anatomical structure hierarchy is added for clarity sake. Note that not all parameter nodes refer to the same volume node, the point is to show that parameter nodes point to some volume node. Also note that parent nodes in the anatomical structure hierarchy are a special case: they do not contain references to their child nodes in the XML tags. However, they do contain pointers to their child nodes in their MRML classes.
Finally, nodes related to the EMSegment module will have EMSegment prepended to the tag name in the graph.
The following list contains all possible template parameters; not all parameters will be tunable by the user during the template building process.
Example Template File
#
# NB: Only the keys are included, not the values.
#
#
# atlas data (intensity and probabiliy maps) are stored in volume nodes
#
<VolumeArchetypeStorage id name>
<VolumeDisplay id name>
<Volume id name storageNodeRef displayNodeRef>
[...]
<VolumeArchetypeStorage id name>
<VolumeDisplay id name>
<Volume id name storageNodeRef displayNodeRef>
#
# template node
#
<EMSegmentTemplate id name globalDisplayNodeRef globalParametersNodeRef
hierarchyNodeRef>
#
# global algorithm parameters
#
<EMSegmentGlobalAlgorithmParameters id name atlasT1NodeRef atlasT2NodeRef
[params...]>
#
# global display parameters
#
<EMSegmentGlobalDisplayParameters id name graphT1NodeRef graphT2NodeRef
[params...]>
<EMSegmentGraphDisplayParameters id name [params...]>
<EMSegmentGraphDisplayParameters id name [params...]>
#
# the hierarchy of anatomical structures is described here
#
<EMSegmentHierarchyNode id name parentNodeRef superClassParametersNodeRef
leafClassParametersNodeRef displayNodeRef>
[...]
#
# node-specific parameters
#
<EMSegmentSuperClassParameters id name cimNorthNodeRef [...] [params...]>
[...]
<EMSegmentLeafClassParameters id name localPriorNodeRef [params...]>
[...]
<EMSegmentClassDisplayParameters id name [params...]>
[...]
#
# class interaction matrices
#
<EMSegmentCIM id name matrix>
[...]
Detailed Description
Atlas Data
- Atlas intensity images (T1 and T2) that are pre-aligned; specified via MRML Volume nodes.
- Spatial prior probability images that are aligned with the atlas intensity images; specified via MRML Volume nodes.
EM Segment Template
The template containes references to global algorithm and display parameters as well as the class hierarchy.
<EMSegmentTemplate id name globalDisplayNodeRef globalParametersNodeRef hierarchyNodeRef>
- MRML node attributes
- Name
- ID
- Core attributes
- GlobalDisplayNodeRef--char*
- GlobalParametersNodeRef--char*
- HierarchyNodeRef--char*
- (I expect this to grow as the GUI develops)
Global EM Algorithm Parameters
Global EM algorithm parameters are stored in a single node.
<EMSegmentGlobalParameters id name atlasT1NodeRef atlasT2NodeRef [params...]>
- MRML node attributes
- Name
- ID
- Core attributes
- Alpha--double (should this be node specific?)
- AlreadyRead--int (internal state variable that should not be included?)
- NumClasses--int (legacy and should be removed?)
- NumberOfEMIterations--int--EMiteration (should this be node specific?)
- NumberOfInputChannels--int--MaxInputChannelDef
- NumberOfMFAIterations--int--MFAiteration (should this be node specific?)
- NumberOfTrainingSamples--int
- SegmentationBoundaryMax--int[3]
- SegmentationBoundaryMin--int[3]
- SmoothingKernelSigma--double--SmSigma (should this be node specific?)
- SmoothingKernelWidth--int--SmWidth (should this be node specific?)
- WorkingDirectory--char*--PrintDir
- Additional core attributes (added by Brad)
- AtlasT1NodeRef--char*
- AtlasT2NodeRef--char*
- RegistratonType--int
- Experimental attributes
- DisableMultiThreading--int
- DisplayProbability--int (change display to print as the others?)
- RegistrationInterpolationType--int
Global EM Display Parameters
Global EM display parameters are stored in a single node.
<EMSegmentGlobalDisplayParameters id name graphT1NodeRef graphT2NodeRef [params...]>
- MRML node attributes
- Name
- ID
- Core attributes
- GraphDisplayT1NodeRef--char*
- GraphDisplayT2NodeRef--char*
- (I expect this to grow as the GUI develops)
Hierarchy of Anatomical Structures
The hierarchy of anatomical structures is defined by a sequence of nodes. The nodes that define the hierarchy are separate from nodes that contain algorithm or display parameters.
<EMSegmentTissueHierarchy id name parentNodeRef superClassParametersNodeRef
leafClassParametersNodeRef disiplayParametersNodeRef>
- MRML node attributes
- Name
- ID
- Core attributes
- ParentNodeRef--char*--null in case of root node
- DisplayParametersNodeRef--char* (for example color in graph)
- SuperClassParametersNodeRef--char*
- LeafClassParametersNodeRef--char*
Node-based Display Parameters
<EMSegmentClassDisplayParameters id name [params...]>
- MRML node attributes
- Name
- ID
- Core attributes
- Color--char*--color associated with this anatomical structure
- I expect that more will be added here as the GUI develops
Generic Node-based EM Algorithm Parameters
There is no MRML node for generic node-based algorithm parameters; rather, these attributes are present in both super-class and leaf-class MRML nodes.
- MRML node attributes
- Name
- ID
- Core attributes
- InputChannelWeights--char*
- LocalPriorVolumeID--char*--LocalPriorName
- LocalPriorWeight--double
- PrintWeights--int
- ClassProbability--double (need more descriptive name)
- Experimental attributes
- ExcludeFromIncompleteEStep--int--ExcludeFromIncompleteEStepFlag
- PrintRegistrationParameters--int
- PrintRegistrationSimilarityMeasure--int
- EmployClassSpecificRegistration--int--RegistrationClassSpecificRegistrationFl\ag
- RegistrationCovariance--double[9]
- RegistrationRotation--double[3]
- RegistrationScale--double[3]
- RegistrationTranslation--double[3]
SuperClass-Specific EM Algorithm Parameters
SuperClass algorithm parameters are stored in a MRML node. Generic node-based attributes are included in this node.
<EMSegmentSuperClassAlgorithmParameters id name [refs to matrix nodes] [params...]>
- MRML node attributes
- Name
- ID
- Core attributes
- InitialBiasFieldPrefix--char* (change to mrml node ID? is this node specific or a global parameter?)
- NumClasses--int
- PredefinedLabelMapPrefix--char* (change to mrml node ID?)
- PrintBias--int
- PrintFrequency--int
- PrintLabelMap--int
- StopEMMaxIteration--int--StopEMMaxIter
- StopEMType--int
- StopEMValue--double
- StopMFAMaxIteration--int--StopMFAMaxIter
- StopMFAType--int
- StopMFAValue--double
- Aditional core attributes (added by Brad)
- CIMMatrixNorthNodeRef--char*
- CIMMatrixSouthNodeRef--char*
- CIMMatrixEastNodeRef--char*
- CIMMatrixWestNodeRef--char*
- CIMMatrixUpNodeRef--char*
- CIMMatrixDownNodeRef--char*
- Experimental attributes
- GenerateBackgroundProbability--int
- ParameterInitSubClass--int
- ParameterSaveToFile--int
- ParameterSetFromFile--int
- PCAShapeModelType--int
- PredefinedLabelID--int (how does this differ from prefix above?)
- PrintEMLabelMapConvergence--int
- PrintEMWeightsConvergence--int
- PrintMFALabelMapConvergence--int
- PrintMFAWeightsConvergence--int
- PrintShapeSimilarityMeasure--int
- RegistrationIndependentSubclassFlag--int (need different name?)
- RegistrationType--int
- StopBiasCalculation--int
LeafClass-Specific EM Algorithm Parameters
LeafClass algorithm parameters are stored in a MRML node. Generic node-based attributes are included in this node.
<EMSegmentLeafClassAlgorithmParameters id name [refs to prior volumes] [params...]>
- MRML node attributes
- Name
- ID
- Core attributes
- Label--int
- LocalPriorWeight--double
- LogCovariance--char*
- LogMean--char*
- PrintQuality--int
- ReferenceStandardFilename--char* (change to mrml node ID?)
- Experimental attributes
- PCALogisticBoundary--double
- PCALogisticMax--double
- PCALogisticMin--double
- PCALogisticSlope--double
- PCAMeanName--char* (a filename?)
- PrintPCA--int
- SamplingLogMean--char*
- SamplingLogCovaraince--char*
Graph Display Parameters
Graph display parameters affect the display of the intensity distribution graphs.
<EMSegmenterGraphDisplay id name [params...]>
- MRML node attributes
- Name
- ID
- Core attributes
- MinimumIntensityValue--int
- MaximumIntensityValue--int
- Scale--int (need more specific name)
Class Interaction Matrix
Six class interaction matrices (CIMs) are included for each parent node in the template. These matrices are named North, South, East, West, Up, and Down. These names correspond to positive and negative cardinal directions within the target image space. The element at row i and column j of the North CIM describes the probability that a voxel with class label i will be found North of a voxel with class label j.
CIMs are represented in the template by EMSegmentCIM nodes.
<EMSegmentCIM id name [matrix entries]>
- MRML node attributes
- Name
- ID
- Core attributes
- Matrix--char*