TubeTK/Documentation/InteractivePDFSegmenter
From KitwarePublic
< TubeTK | Documentation
Jump to navigationJump to search
Revision as of 21:42, 17 January 2011 by Danielle.pace (talk | contribs)
Overview
The interactive PDF segmenter performs image segmentation based on a rough segmentation provided by the user. This rough segmentation can be "painted" by the user within 3D Slicer, and can be refined iteratively after visualizing the segmentation results.
The segmentation steps proceed as follows:
- Using the intensity statistics within the rough segmentation, calculate the class probabilities for each pixel in the image to be segmented
- Smooth the probability map
- Classify each pixel to the class with the highest probability: now every pixel has a label
- Analyze connected components relative to the rough segmentation: retain only those classified pixels that are connected to a pixel of the same class in the rough segmentation
- Perform holefilling on the label map
- Perform erosion on the label map
- Perform connected components analysis again: should now have well connected regions
- Perform dilation on the label map
Running the Interactive PDF Segmenter
The interactive PDF segmenter is to be used with 3D Slicer (see the TubeTK with Slicer page for setup instructions).
Its implementation can be found in TubeTK/Applications/SlicerModules/InteractivePDFSegmenter.py.
Parameters
- Input Volume 1: The image to be segmented
- Input Volumes 2 and 3 (optional): TODO
- Output Volume: The segmentation results
- Label Map: A rough initial segmentation provided as input to the algorithm
- Void Id: Value that specifies nothing in the label map
- Erosion Radius: TODO
- Hole Fill Iterations: TODO
- False Positive Ratio: TODO
- Probability Smoothing Standard Deviations: TODO
- Draft Mode: TODO
- Reclassify Object Mask: Whether or not to TODO
- Reclassify Not Object Mask: Whether or not to TODO
Tutorial
Tutorial Data
- In this example, we will attempt to segment gray matter, white matter and cerebrospinal fluid (CSF)
- The 2D brain image used in this example is distributed as part of Slicer4, and is located in Slicer4/Testing/Data/Input/brainSliceDOUBLE.mha
Tutorial Steps
- Load brainSliceDOUBLE.mha:
- File -> Add Volume
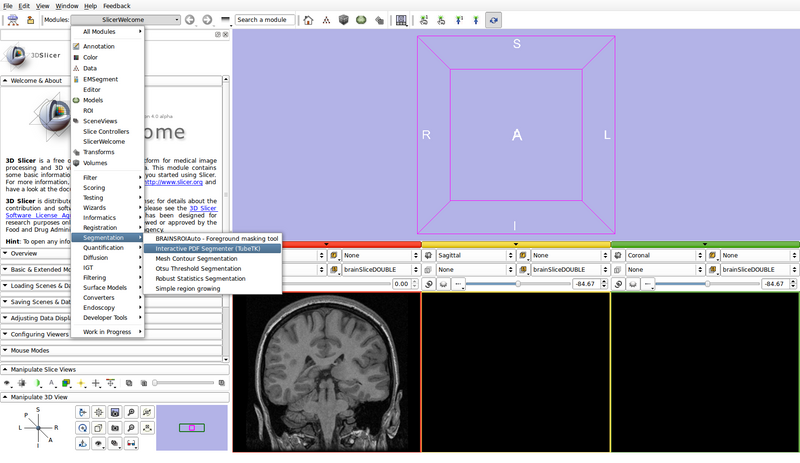
- Open the Interactive PDF Segmenter module
- Set the input and output volumes
- Input Volume 1: brainSliceDOUBLE
- Input Volume 2 (optional): None
- Input Volume 3 (optional): None
- Output Volume: Create new volume
- Creates a new volume called "Volume": this will hold the final segmentation
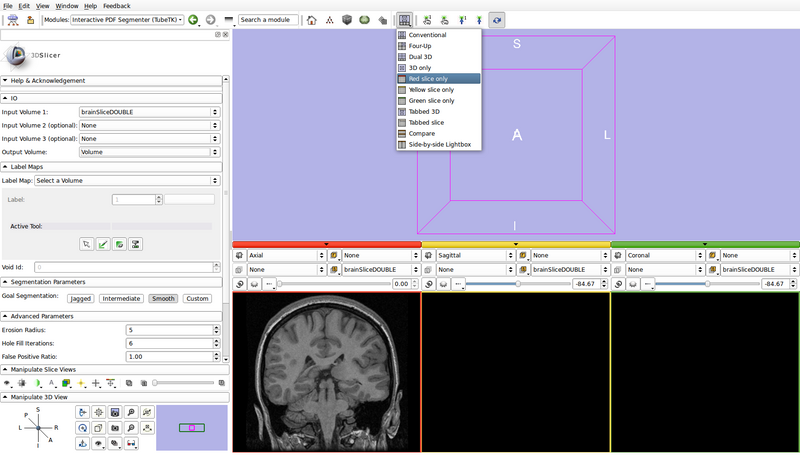
- Set the view layout to "Red slice only"
- Create the label map roughly outlining the structures to be segmented and the background
- Label Map: Create new volume
- Creates a new volume called "Volume1": this will hold the input label map
- A small version of Slicer's Editor module is embedded into the Interactive PDF Segmenter
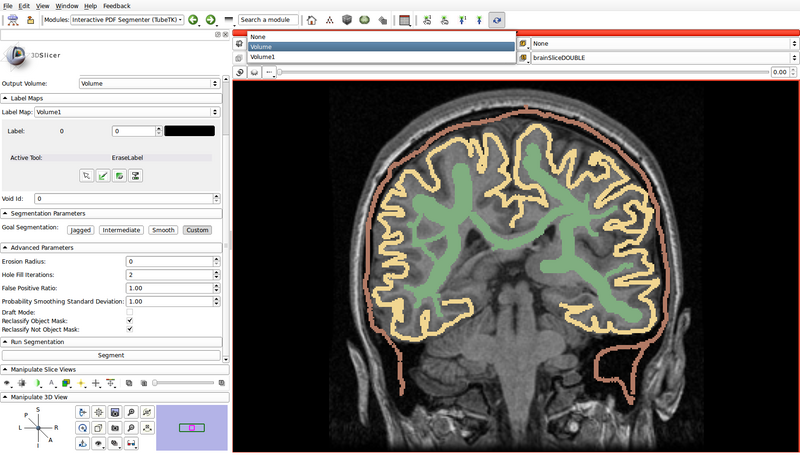
- Create a label map roughly outlining the structures of interest and the background by using the paint, erase and threshold functions
- Click on the "Paint" button to begin editing
- Use a different label color for each structure of interest: choose the label by clicking the colored push button
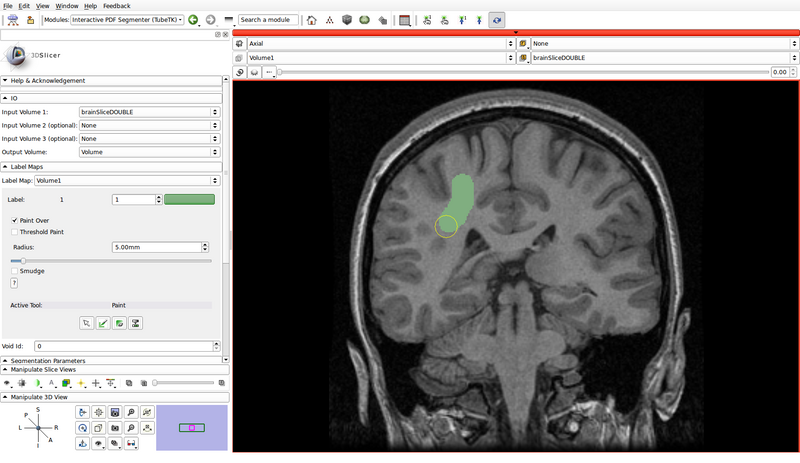
- Create a label for the background as well
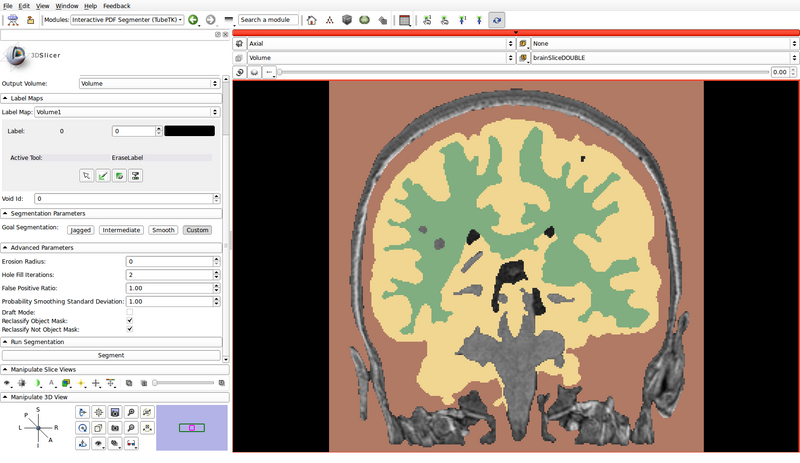
- In the following example, green represents gray matter, yellow represents white matter, and brown represents the cerebrospinal fluid (CSF):
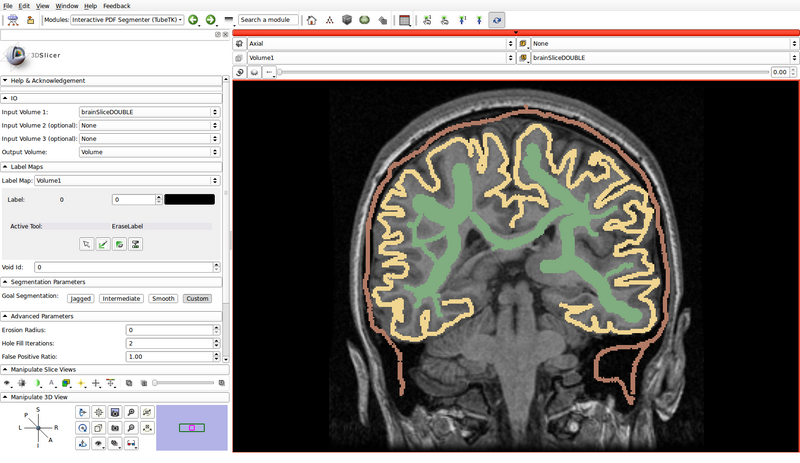
- Void Id: 0
- Label Map: Create new volume
- Specify the segmentation parameters
- The goal segmentation buttons provide example parameters depending on whether the shapes of the expected segmentations are jagged, smooth or intermediate
- In this example, we will use custom parameters
- Erosion Radius: 0
- Hole Fill Iterations: 2
- False Positive Ratio: 1.00
- Probability Smoothing Standard Deviation: 1.00
- Draft Mode: Off
- Reclassify Object Mask: On
- Reclassify Not Object Mask: On
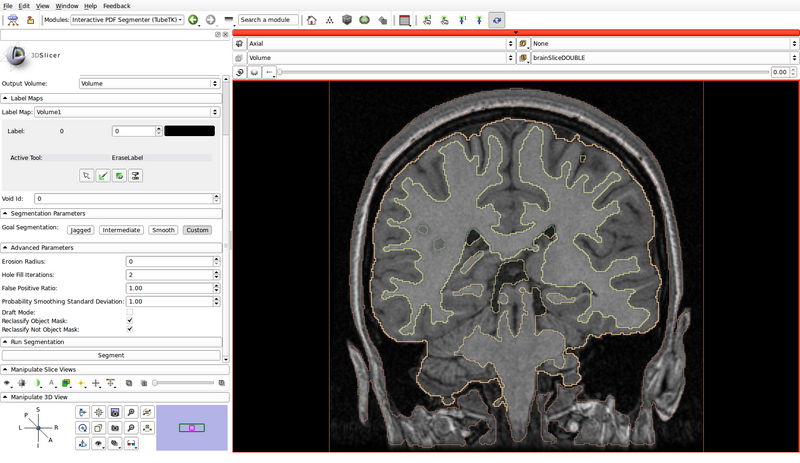
- Run the segmentation